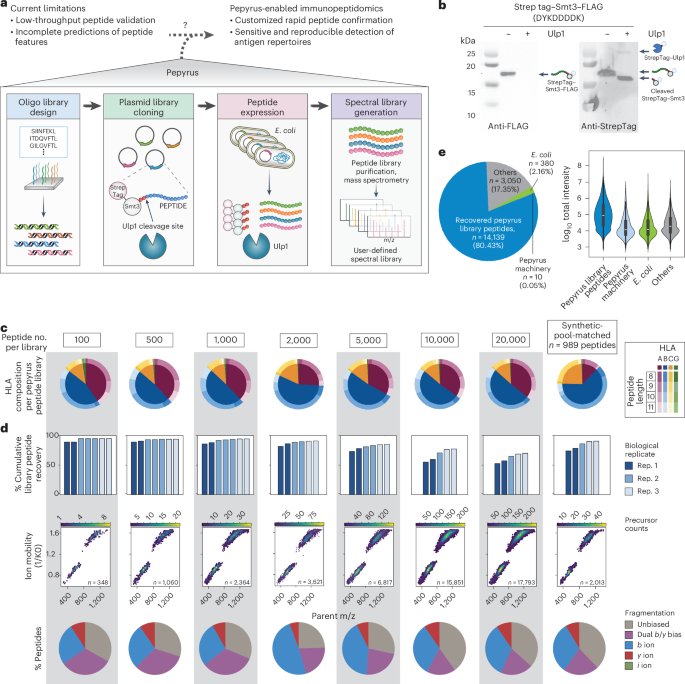
Sensitive detection of cancer antigens enabled by user-defined peptide libraries – Nature Biotechnology
Haen, S. P., Löffler, M. W., Rammensee, H.-G. & Brossart, P. Towards new horizons: characterization, classification and implications of the tumour antigenic repertoire. Nat. Rev. Clin. Oncol. 17, 595–610 (2020).
Google Scholar
Sensi, M. & Anichini, A. Unique tumor antigens: evidence for immune control of genome integrity and immunogenic targets for T cell-mediated patient-specific immunotherapy. Clin. Cancer Res. 12, 5023–5032 (2006).
Google Scholar
Sellars, M. C., Wu, C. J. & Fritsch, E. F. Cancer vaccines: building a bridge over troubled waters. Cell 185, 2770–2788 (2022).
Google Scholar
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Google Scholar
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Google Scholar
Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019).
Google Scholar
Rojas, L. A. et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 618, 144–150 (2023).
Google Scholar
Hong, D. S. et al. Autologous T cell therapy for MAGE-A4+ solid cancers in HLA-A*02+ patients: a phase 1 trial. Nat. Med. 29, 104–114 (2023).
Google Scholar
Parkhurst, M. et al. Adoptive transfer of personalized neoantigen-reactive TCR-transduced T cells in metastatic colorectal cancer: phase 2 trial interim results. Nat. Med. 30, 2586–2595 (2024).
Google Scholar
Chong, C., Coukos, G. & Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol. 40, 175–188 (2022).
Google Scholar
Sarkizova, S. et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 38, 199–209 (2020).
Google Scholar
Michalski, A., Cox, J. & Mann, M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC–MS/MS. J. Proteome Res. 10, 1785–1793 (2011).
Google Scholar
Granados, D. P. et al. Impact of genomic polymorphisms on the repertoire of human MHC class I-associated peptides. Nat. Commun. 5, 3600 (2014).
Google Scholar
van der Lee, D. I. et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J. Clin. Invest. 129, 774–785 (2019).
Google Scholar
Pak, H. et al. Sensitive immunopeptidomics by leveraging available large-scale multi-HLA spectral libraries, data-independent acquisition, and MS/MS prediction. Mol. Cell. Proteomics 20, 100080 (2021).
Google Scholar
Gabriel, W., Picciani, M., The, M. & Wilhelm, M. Deep learning-assisted analysis of immunopeptidomics data. Methods Mol. Biol. 2758, 457–483 (2024).
Google Scholar
Adams, C. et al. Fragment ion intensity prediction improves the identification rate of non-tryptic peptides in timsTOF. Nat. Commun. 15, 3956 (2024).
Google Scholar
Oliinyk, D. et al. diaPASEF analysis for HLA-I peptides enables quantification of common cancer neoantigens. Mol. Cell Proteomics 24, 100938 (2025).
Google Scholar
Gessulat, S. et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 16, 509–518 (2019).
Google Scholar
Bouwmeester, R., Gabriels, R., Hulstaert, N., Martens, L. & Degroeve, S. DeepLC can predict retention times for peptides that carry as-yet unseen modifications. Nat. Methods 18, 1363–1369 (2021).
Google Scholar
Zeng, W.-F. et al. AlphaPeptDeep: a modular deep learning framework to predict peptide properties for proteomics. Nat. Commun. 13, 7238 (2022).
Google Scholar
C Silva, A. S., Bouwmeester, R., Martens, L. & Degroeve, S. Accurate peptide fragmentation predictions allow data driven approaches to replace and improve upon proteomics search engine scoring functions. Bioinformatics 35, 5243–5248 (2019).
Google Scholar
Degroeve, S. & Martens, L. MS2PIP: a tool for MS/MS peak intensity prediction. Bioinformatics 29, 3199–3203 (2013).
Google Scholar
Wilhelm, M. et al. Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics. Nat. Commun. 12, 3346 (2021).
Google Scholar
Declercq, A. et al. TIMS2Rescore: a data dependent acquisition-parallel accumulation and serial fragmentation-optimized data-driven rescoring pipeline based on MS2Rescore. J. Proteome Res. 24, 1067–1076 (2025).
Google Scholar
Strauss, M. T. et al. AlphaPept: a modern and open framework for MS-based proteomics. Nat. Commun. 15, 2168 (2024).
Google Scholar
Lee, C.-D. et al. An improved SUMO fusion protein system for effective production of native proteins. Protein Sci. 17, 1241–1248 (2008).
Google Scholar
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Google Scholar
Roepstorff, P. & Fohlman, J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601 (1984).
Google Scholar
Zubarev, R. A. & Makarov, A. Orbitrap mass spectrometry. Anal. Chem. 85, 5288–5296 (2013).
Google Scholar
Prensner, J. R. et al. What can Ribo-Seq, immunopeptidomics, and proteomics tell us about the noncanonical proteome? Mol. Cell. Proteomics 22, 100631 (2023).
Google Scholar
Skiadopoulou, D. et al. Retention time and fragmentation predictors increase confidence in identification of common variant peptides. J. Proteome Res. 22, 3190–3199 (2023).
Google Scholar
Frewen, B. E., Merrihew, G. E., Wu, C. C., Noble, W. S. & MacCoss, M. J. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 78, 5678–5684 (2006).
Google Scholar
Lam, H. et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 (2007).
Google Scholar
Yang, X. et al. Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides. Nature 612, 771–777 (2022).
Google Scholar
Phulphagar, K. M. et al. Sensitive, high-throughput HLA-I and HLA-II immunopeptidomics using parallel accumulation–serial fragmentation mass spectrometry. Mol. Cell. Proteomics 22, 100563 (2023).
Google Scholar
Malakhov, M. P. et al. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics 5, 75–86 (2004).
Google Scholar
Kapust, R. B., Tözsér, J., Copeland, T. D. & Waugh, D. S. The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 294, 949–955 (2002).
Google Scholar
Chang, J. Y. Thrombin specificity. Requirement for apolar amino acids adjacent to the thrombin cleavage site of polypeptide substrate. Eur. J. Biochem. 151, 217–224 (1985).
Google Scholar
Gomez-Zepeda, D. et al. Thunder-DDA-PASEF enables high-coverage immunopeptidomics and is boosted by MS2Rescore with MS2PIP timsTOF fragmentation prediction model. Nat. Commun. 15, 2288 (2024).
Google Scholar
Käll, L., Storey, J. D., MacCoss, M. J. & Noble, W. S. Posterior error probabilities and false discovery rates: two sides of the same coin. J. Proteome Res. 7, 40–44 (2008).
Google Scholar
Oliveira, G. et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 596, 119–125 (2021).
Google Scholar
Oliveira, G. et al. Landscape of helper and regulatory antitumour CD4+ T cells in melanoma. Nature 605, 532–538 (2022).
Google Scholar
Klaeger, S. et al. Optimized liquid and gas phase fractionation increases HLA-peptidome coverage for primary cell and tissue samples. Mol. Cell. Proteomics 20, 100133 (2021).
Google Scholar
Hodis, E. et al. A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012).
Google Scholar
Ouspenskaia, T. et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat. Biotechnol. 40, 209–217 (2022).
Google Scholar
Eisenberg, G. et al. Transcutaneous immunization with hydrophilic recombinant gp100 protein induces antigen-specific cellular immune response. Cell. Immunol. 266, 98–103 (2010).
Google Scholar
Kawakami, Y. et al. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J. Immunol. 154, 3961–3968 (1995).
Google Scholar
Salgaller, M. L. et al. Recognition of multiple epitopes in the human melanoma antigen gp100 by peripheral blood lymphocytes stimulated in vitro with synthetic peptides. Cancer Res. 55, 4972–4979 (1995).
Google Scholar
Bruderer, R. et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics 14, 1400–1410 (2015).
Google Scholar
Kawakami, Y. et al. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J. Exp. Med. 180, 347–352 (1994).
Google Scholar
Braun, D. A. et al. A neoantigen vaccine generates antitumour immunity in renal cell carcinoma. Nature 639, 474–482 (2025).
Google Scholar
Jiang, Q. et al. HIF regulates multiple translated endogenous retroviruses: implications for cancer immunotherapy. Cell 188, 1807–1827 (2025).
Google Scholar
Takahashi, Y. et al. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J. Clin. Invest. 118, 1099–1109 (2008).
Google Scholar
Bonaventura, P. et al. Identification of shared tumor epitopes from endogenous retroviruses inducing high-avidity cytotoxic T cells for cancer immunotherapy. Sci. Adv. 8, eabj3671 (2022).
Google Scholar
Saini, S. K. et al. Human endogenous retroviruses form a reservoir of T cell targets in hematological cancers. Nat. Commun. 11, 5660 (2020).
Google Scholar
Smith, C. C. et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Invest. 128, 4804–4820 (2018).
Google Scholar
Bassani-Sternberg, M. et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404 (2016).
Google Scholar
Yi, X. et al. caAtlas: an immunopeptidome atlas of human cancer. iScience 24, 103107 (2021).
Google Scholar
Kraemer, A. I. et al. The immunopeptidome landscape associated with T cell infiltration, inflammation and immune editing in lung cancer. Nat. Cancer 4, 608–628 (2023).
Google Scholar
Ferreira, H. J. et al. Immunopeptidomics-based identification of naturally presented non-canonical circRNA-derived peptides. Nat. Commun. 15, 2357 (2024).
Google Scholar
Liao, H. et al. MARS an improved de novo peptide candidate selection method for non-canonical antigen target discovery in cancer. Nat. Commun. 15, 661 (2024).
Google Scholar
Minegishi, Y. et al. Differential ion mobility mass spectrometry in immunopeptidomics identifies neoantigens carrying colorectal cancer driver mutations. Commun. Biol. 5, 831 (2022).
Google Scholar
Hoenisch Gravel, N. et al. TOFIMS mass spectrometry-based immunopeptidomics refines tumor antigen identification. Nat. Commun. 14, 7472 (2023).
Google Scholar
Wahle, M. et al. IMBAS-MS discovers organ-specific HLA peptide patterns in plasma. Mol. Cell. Proteomics 23, 100689 (2024).
Google Scholar
Sahin, U. & Türeci, Ö Personalized vaccines for cancer immunotherapy. Science 359, 1355–1360 (2018).
Google Scholar
Huber, F. et al. A comprehensive proteogenomic pipeline for neoantigen discovery to advance personalized cancer immunotherapy. Nat. Biotechnol. 43, 1360–1372 (2025).
Google Scholar
Bruno, P. M. et al. High-throughput, targeted MHC class I immunopeptidomics using a functional genetics screening platform. Nat. Biotechnol. 41, 980–992 (2023).
Google Scholar
Chong, C. et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat. Commun. 11, 1293 (2020).
Google Scholar
Cormican, J. A., Soh, W. T., Mishto, M. & Liepe, J. iBench: a ground truth approach for advanced validation of mass spectrometry identification method. Proteomics 23, e2200271 (2023).
Google Scholar
Parker, R. et al. The choice of search engine affects sequencing depth and HLA class I allele-specific peptide repertoires. Mol. Cell. Proteomics 20, 100124 (2021).
Google Scholar
Harper, S. & Speicher, D. W. Purification of proteins fused to glutathione S-transferase. Methods Mol. Biol. 681, 259–280 (2011).
Google Scholar
Zhu, K. et al. Cleavage of fusion proteins on the affinity resins using the TEV protease variant. Protein Expr. Purif. 131, 27–33 (2017).
Google Scholar
Lau, Y.-T. K. et al. Discovery and engineering of enhanced SUMO protease enzymes. J. Biol. Chem. 293, 13224–13233 (2018).
Google Scholar
Alspach, E. et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–701 (2019).
Google Scholar
Abelin, J. G. et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017).
Google Scholar
Chen, B. et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 37, 1332–1343 (2019).
Google Scholar
Deutsch, E. W. et al. High-quality peptide evidence for annotating non-canonical open reading frames as human proteins. Preprint at bioRxiv https://doi.org/10.1101/2024.09.09.612016 (2024).
Soufi, B. & Macek, B. Stable isotope labeling by amino acids applied to bacterial cell culture. Methods Mol. Biol. 1188, 9–22 (2014).
Google Scholar
Park, H.-S. et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–1154 (2011).
Google Scholar
Fan, C., Ip, K. & Söll, D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 590, 3040–3047 (2016).
Google Scholar
Ojima-Kato, T., Nagai, S. & Nakano, H. N-terminal SKIK peptide tag markedly improves expression of difficult-to-express proteins in Escherichia coli and Saccharomyces cerevisiae. J. Biosci. Bioeng. 123, 540–546 (2017).
Google Scholar
Berman, H. M. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Google Scholar
Yu, F. et al. Fast quantitative analysis of timsTOF PASEF data with MSFragger and IonQuant. Mol. Cell. Proteomics 19, 1575–1585 (2020).
Google Scholar
da Veiga Leprevost, F. et al. Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nat. Methods 17, 869–870 (2020).
Google Scholar
Yang, K. L. et al. MSBooster: improving peptide identification rates using deep learning-based features. Nat. Commun. 14, 4539 (2023).
Google Scholar
Yu, F., Haynes, S. E. & Nesvizhskii, A. I. IonQuant enables accurate and sensitive label-free quantification with FDR-controlled match-between-runs. Mol. Cell. Proteomics 20, 100077 (2021).
Google Scholar
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Google Scholar
Kong, A. T., Leprevost, F. V., Avtonomov, D. M., Mellacheruvu, D. & Nesvizhskii, A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017).
Google Scholar
Li, K., Teo, G. C., Yang, K. L., Yu, F. & Nesvizhskii, A. I. diaTracer enables spectrum-centric analysis of diaPASEF proteomics data. Nat. Commun. 16, 95 (2025).
Google Scholar
Tareen, A. & Kinney, J. B. Logomaker: beautiful sequence logos in Python. Bioinformatics 36, 2272–2274 (2020).
Google Scholar
Henikoff, S. & Henikoff, J. G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA 89, 10915–10919 (1992).
Google Scholar
Bremel, R. D. & Homan, E. J. An integrated approach to epitope analysis I: dimensional reduction, visualization and prediction of MHC binding using amino acid principal components and regression approaches. Immunome Res. 6, 7 (2010).
Google Scholar
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3, 861 (2018).
Google Scholar
Ester, M. Density-based clustering. In Data Clustering (eds Aggarwal, C. C. & Reddy, C. K.) (Chapman and Hall/CRC, 2018).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Google Scholar